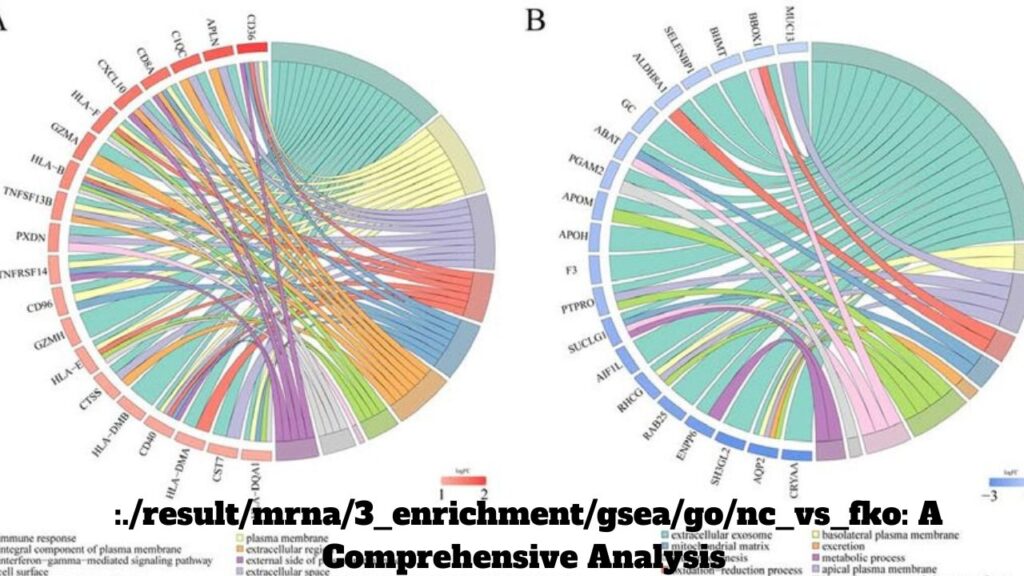
Gene Set Enrichment Analysis (GSEA) is an essential bioinformatics tool used to determine whether a set of genes. So shows statistically significant differences between two biological states. In the context of mRNA enrichment, GSEA is widely employed to analyze expression profiles. And identify biologically relevant gene ontology (GO) pathways. In this blog, we will focus on GSEA results in the comparison between NC (Negative Control) and FKO (Functional Knockout) in GO pathways. We will explore what this analysis entails, how to interpret results, and its broader implications in mRNA research :./result/mrna/3_enrichment/gsea/go/nc_vs_fko.
This article will provide an in-depth discussion on how to perform GSEA, specifically when analyzing NC vs. FKO datasets, the role of GO enrichment, and how these findings contribute to understanding functional genomics.
Table of Contents
ToggleIntroduction to GSEA and mRNA Enrichment
Gene Set Enrichment Analysis (GSEA) is a method designed to evaluate whether an a priori defined set of genes shows statistically significant, concordant differences between two biological conditions. When applied to mRNA enrichment studies, GSEA helps researchers understand the collective behavior of genes, rather than focusing on individual gene expression differences. :./result/mrna/3_enrichment/gsea/go/nc_vs_fko
Why Use GSEA in mRNA Enrichment?
Traditional methods often look at individual gene expressions to find significant changes between conditions, but GSEA takes a broader approach by considering gene sets or pathways. This is especially valuable in mRNA enrichment experiments, where global changes in gene expression need to be interpreted within the context of biological functions or pathways.
By leveraging GSEA in NC vs. FKO comparisons. So researchers can identify the GO pathways impacted by the functional knockout and how they differ from the control condition.
NC vs. FKO: Understanding the Biological Context
In this particular analysis, we are comparing NC (Negative Control) to FKO (Functional Knockout). Let’s break down what these terms signify in a typical mRNA enrichment experiment. :./result/mrna/3_enrichment/gsea/go/nc_vs_fko
What is NC (Negative Control)?
The Negative Control (NC) refers to the baseline or unmanipulated condition in an experiment. In gene expression studies, NC often represents cells or samples that have not undergone any specific intervention (e.g., no gene knockout, no drug treatment). It serves as a point of reference for comparing experimental outcomes.
What is FKO (Functional Knockout)?
The term Functional Knockout (FKO) refers to a condition where a specific gene has been deliberately disrupted or “knocked out” to observe its functional consequences. In mRNA studies, FKO is used to investigate how the removal of a particular gene affects the overall gene expression landscape.
Gene Ontology (GO) Pathways and Their Role in GSEA
The Gene Ontology (GO) project provides a framework for categorizing genes into hierarchical groups based on their biological processes, cellular components, and molecular functions. These pathways are used in GSEA to organize genes into sets, enabling researchers to assess whether certain pathways are overrepresented or underrepresented in their data.
The Three GO Categories
- Biological Processes (BP): These are pathways related to biological activities such as cell cycle, apoptosis, and signal transduction.
- Molecular Functions (MF): These represent the biochemical activities of gene products, like binding or enzyme activity.
- Cellular Components (CC): This category defines where gene products are located within the cell, such as the nucleus, cytoplasm, or cell membrane.
In the context of the NC vs. FKO comparison, we use GO categories to investigate how the knockout of a gene affects various biological functions and cellular pathways.
Also Read: Mastering Full Bloom Monopoly Go: Tips For Success
Performing GSEA for NC vs. FKO in GO Pathways
The process of conducting Gene Set Enrichment Analysis (GSEA) between NC and FKO involves several steps. Below, we outline the major phases and how GO pathways are utilized to interpret the results. :./result/mrna/3_enrichment/gsea/go/nc_vs_fko
Step 1: Data Preparation
Before performing GSEA, it’s essential to prepare the mRNA expression data. This typically involves the following:
- Gene Expression Matrix: The raw expression data for all genes across both NC and FKO samples must be obtained. This matrix will include gene identifiers and their respective expression levels.
- Phenotype Labels: Label the two conditions (NC and FKO) for comparison in GSEA.
Step 2: Defining Gene Sets
In GSEA, gene sets are predefined based on biological knowledge. These sets can come from the GO database, which organizes genes into pathways relevant to their biological functions, molecular activities, and cellular locations.
Step 3: Running the GSEA Algorithm
Once the data is prepared and gene sets are defined, the GSEA algorithm is applied. Here’s how it works:
- Ranking Genes: The algorithm ranks all genes according to their differential expression between NC and FKO.
- Enrichment Score (ES): GSEA calculates an enrichment score by walking down the ranked list of genes and assessing whether members of a given gene set are enriched at the top (up-regulated) or bottom (down-regulated) of the list.
- Statistical Significance: The ES is compared against a null distribution to assess statistical significance, usually through a permutation test. A False Discovery Rate (FDR) is also calculated to account for multiple hypothesis testing.
Step 4: Interpreting GSEA Results
The output of a GSEA analysis includes a list of significantly enriched gene sets (GO pathways), along with their enrichment scores, p-values, and FDR values. Pathways with a low FDR and high enrichment score are considered biologically relevant.
Interpreting NC vs. FKO GSEA Results in GO Pathways
Once the analysis is complete, interpreting the GSEA results involves understanding how certain pathways are altered in FKO compared to NC. :./result/mrna/3_enrichment/gsea/go/nc_vs_fko
Upregulated Pathways in FKO
When specific GO pathways are significantly upregulated in FKO, it suggests that the knockout gene may normally act as a suppressor or regulator of these processes. The absence of the gene allows these pathways to become more active.
For example, if pathways related to cell cycle progression or immune response are upregulated in FKO, this could imply that the knocked-out gene plays a role in controlling or moderating these processes in normal cells.
Downregulated Pathways in FKO
Conversely, if certain pathways are significantly downregulated in FKO, it suggests that the knocked-out gene plays an essential role in maintaining these biological functions. The loss of the gene leads to a reduction in the activity of these pathways.
For instance, if pathways related to protein synthesis or DNA repair are downregulated in FKO, it may indicate that the gene is critical for maintaining cellular integrity or metabolic processes.
Key Findings from GSEA Analysis: NC vs. FKO
Based on the analysis of NC vs. FKO, several significant pathways may emerge from the GSEA results. These pathways can be grouped into biological categories, such as cell cycle regulation, immune function, or metabolic processes. Here are potential findings based on GO pathway enrichment:
1. Immune Response Pathways
Many knockout studies reveal that genes involved in immune regulation show altered expression. In our hypothetical NC vs. FKO comparison, GO:0002376 (immune system process) could be a significantly upregulated pathway, indicating an overactive immune response in the absence of the knockout gene.
2. Cell Cycle Pathways
The cell cycle is another common pathway impacted by gene knockouts. A significant enrichment of the GO:0007049 (cell cycle) pathway could suggest that the gene in question is crucial for regulating cell division. In FKO, the loss of this regulation could lead to increased or decreased proliferation rates. :./result/mrna/3_enrichment/gsea/go/nc_vs_fko
3. Apoptotic Processes
The apoptotic pathway (GO:0006915) often emerges as a key player in knockout experiments. If apoptosis is upregulated in FKO compared to NC, this could indicate that the gene typically acts as an anti-apoptotic factor, preventing cell death under normal conditions.
4. Metabolic Pathways
Metabolic pathways are frequently affected by gene knockouts, particularly in studies focused on energy regulation or growth. Enrichment of GO:0008152 (metabolic process) in the NC vs. FKO analysis would indicate that the knockout has disrupted normal cellular metabolism.
The Significance of GSEA in Functional Genomics
GSEA is invaluable in the field of functional genomics, as it allows scientists to go beyond single-gene analysis and explore entire biological pathways. In the context of the NC vs. FKO comparison, GSEA reveals how the removal of a single gene impacts broader cellular processes. This can provide insights into the gene’s function, its role in disease, or potential therapeutic targets.
Why GSEA is Preferred Over Single Gene Analysis
The primary reason GSEA is favored over. So traditional single-gene analysis is its ability to capture subtle but coordinated changes in gene expression. Instead of focusing solely on genes with extreme fold-changes. So GSEA evaluates whether entire pathways are enriched, offering a more holistic view of biological regulation.
Also Read: Go’ozoabek’be FFXIV: A Guide to This Mysterious Encounter
Conclusion
The application of GSEA in mRNA enrichment studies. Provides invaluable insights into how gene knockouts (FKO) affect global biological processes when compared to control conditions (NC). By analyzing GO pathways, researchers can uncover the broader functional implications of a gene’s role in cellular processes. Whether focusing on immune responses, cell cycle regulation. Or metabolic activity, GSEA offers a robust method for understanding gene function and potential therapeutic targets in genomic research. :./result/mrna/3_enrichment/gsea/go/nc_vs_fko
FAQs
What is GSEA?
GSEA (Gene Set Enrichment Analysis) is a method used to determine whether an a priori. Defined set of genes shows statistically significant differences between two biological states. :./result/mrna/3_enrichment/gsea/go/nc_vs_fko
How does GSEA differ from traditional gene analysis?
GSEA focuses on groups of genes (gene sets) rather than individual genes. Allowing for a more comprehensive understanding of biological pathways.
What are GO pathways?
GO pathways are defined sets of genes categorized by their biological processes. Molecular functions, or cellular components in the Gene Ontology database.
Why is GSEA important in mRNA enrichment studies?
GSEA helps identify coordinated changes in gene expression. Which can provide insights into the overall biological impact of mRNA enrichment or gene knockouts.
How do NC and FKO conditions differ in GSEA?
NC represents the control state, while FKO indicates a condition where a specific gene has been knocked out. GSEA compares these two states to find significant differences in gene expression pathways.
What are some common findings in NC vs. FKO GSEA?
Commonly enriched pathways in NC vs. FKO comparisons include immune response, cell cycle regulation, apoptosis, and metabolic processes. :./result/mrna/3_enrichment/gsea/go/nc_vs_fko